research topics
Protein language models for understanding
and guiding evolution
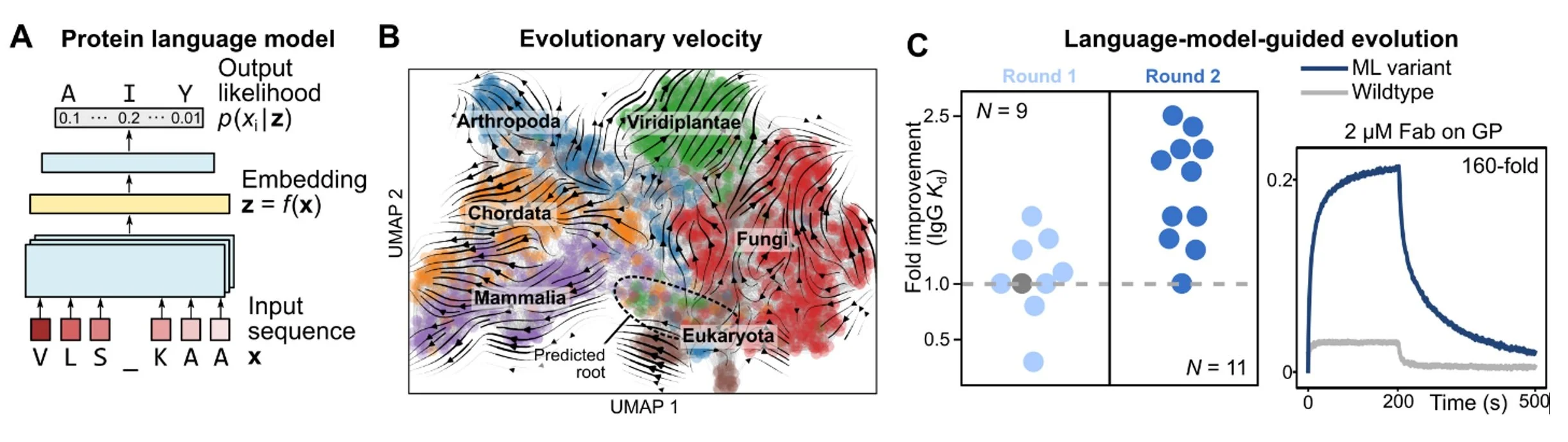
Nature is the most powerful protein engineer, and evolution is nature’s design process. We have been using machine learning algorithms to better understand the rules of natural protein evolution, with the ultimate goal of designing better proteins. We leverage sophisticated algorithms called neural language models that can learn evolutionary patterns from millions of natural protein sequences (Fig. 11A).
To demonstrate that language models can learn evolution, we first apply them to the task of predicting evolutionary dynamics, which has diverse applications that range from tracing the progression of viral outbreaks to understanding the history of life on earth. Our main conceptual advance is to reconstruct a global landscape of protein evolution through local evolutionary predictions that we refer to as evolutionary velocity (evo-velocity), which predicts the direction of evolution in landscapes of viral proteins evolving over years and of eukaryotic proteins evolving over geologic eons (Fig. 11B).
The same ideas can also be used to design new proteins. By testing mutations with higher language-model likelihood than wildtype (i.e., with positive “evolutionary velocity”), we find a surprisingly large number of variants with improved fitness. This enables highly efficient, machine-learning-guided antibody affinity maturation against diverse viral antigens, without providing the model with knowledge of the antigen, protein structure, or task-specific training data (Fig. 11C), and may also inform the design of other types of proteins as well.
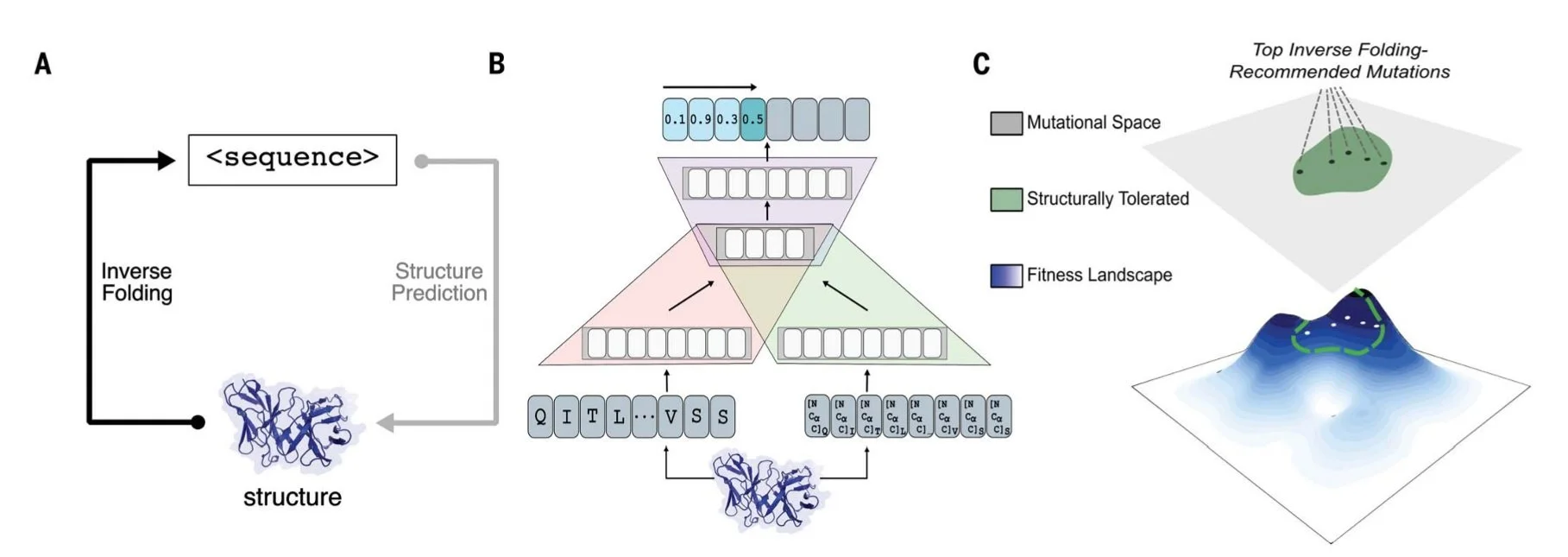
While our work established the effectiveness of language models to guide evolution using sequence alone, a protein's function is inherently linked to its underlying structure. To improve predictive capabilities, we incorporate structural information with a structure-informed language model and demonstrate substantial gains across protein families and definitions of protein fitness. Importantly, we show that using only protein backbone coordinates and no side-chain information is sufficient to learn principles of binding that generalize to protein complexes and enable antibody engineering with unprecedented efficiency. Together, these results lay the groundwork for more potent and resilient therapeutic design in completely unsupervised settings, or even in the absence of task-specific training data (Fig 12).
Figure 11. (A) A language model learns the likelihood of an amino-acid residue occurring within a sequence context. When trained on millions of protein sequences, a large language model can capture the evolutionary “rules” governing which combinations of amino acids are more likely to appear with each other. (B) An evolutionary landscape can be approximated by a composition of local evolutionary predictions made by a language model, which we refer to as evo-velocity. Evo-velocity can predict the directionality of evolution; for example, evo-velocity analysis of the landscape of cytochrome c sequences recovers the order of various taxonomic classes in the fossil record. (C) Language models can also help guide evolution. By selecting mutations with higher language-model-likelihood than wildtype, we find many that also improve protein function under specific notions of fitness. In particular, we use language models to guide structure- and antigen-agnostic antibody affinity maturation against diverse viral antigens.
Figure 12. We utilize a structure-informed language model to consider the inverse folding task of predicting amino acid sequence from protein backbone structure and indirectly explore protein function fitness landscapes. For protein engineering tasks, this allows us to constrain directed evolution exploration to regimes where the protein fold is preserved. We demonstrate the effectiveness of this method to evolve antibodies for improved potency and resilience against viral escape variants.
PUBLIcation highlights
Unsupervised evolution of protein and antibody complexes with a structure-informed language model. (PDF) (Science)
Varun R. Shanker, Theodora U. J. Bruun, Brian L. Hie, Peter S. Kim. Science 385, 46-53 (2024). doi: 10.1126/science.adk8946.
Supplementary Materials
Efficient evolution of human antibodies from general protein language models. (PDF) (Nat. Bio.)
Brian L. Hie, Varun R. Shanker, Duo Xu, Theodora U. J. Bruun, Payton A. Weidenbacher, Shaogeng Tang, Wesley Wu, John E. Pak, Peter S. Kim. Nature Biotechnology (2023) doi.org/10.1038/s41587-023-01763-2.
Evolutionary velocity with protein language models predicts evolutionary dynamics of diverse proteins. (PDF) (Cell Systems)
Brian L.Hie, Kevin K. Yang, Peter S.Kim. Cell Systems (2022).